Normalization.jl
This package allows you to easily normalize an array over any combination of dimensions, with a bunch of methods (z-score, sigmoid, centering, minmax, etc.) and modifiers (robust, mixed, NaN-safe).
Usage
Each normalization method is a subtype of AbstractNormalization. Each AbstractNormalization subtype has its own estimators and forward methods that define how parameters are calculated and the normalization formula. Each AbstractNormalization instance contains the concrete parameter values for a normalization, fit to a given input array.
You can work with AbstractNormalizations as either types or instances. The type approach is useful for concise code, whereas the instance approach is useful for performant mutations. In the examples below we use the ZScore normalization, but the same syntax applies to all Normalizations.
Fit to a type
X = randn(100, 10)
N = fit(ZScore, X; dims=nothing) # eltype inferred from X
N = fit(ZScore{Float32}, X; dims=nothing) # eltype set to Float32
N isa AbstractNormalization && N isa ZScore # Returns a concrete AbstractNormalizationFit to an instance
X = randn(100, 10)
N = ZScore{Float64}(; dims=2) # Initializes with empty parameters
N isa AbstractNormalization && N isa ZScore # Returns a concrete AbstractNormalization
!isfit(N)
fit!(N, X; dims=1) # Fit normalization in-place, and update the `dims`
Normalization.dims(N) == 1Normalization and denormalization
With a fit normalization, there are two approaches to normalizing data: in-place and out-of-place.
_X = copy(X)
normalize!(_X, N) # Normalizes in-place, updating _X
Y = normalize(X, N) # Normalizes out-of-place, returning a new array
normalize(X, ZScore; dims=1) # For convenience, fits and then normalizesFor most normalizations, there is a corresponding denormalization that transforms data to the original space.
Z = denormalize(Y, N) # Denormalizes out-of-place, returning a new array
Z ≈ X
denormalize!(Y, N) # Denormalizes in-place, updating YBoth syntaxes allow you to specify the dimensions to normalize over. For example, to normalize each 2D slice (i.e. iterating over the 3rd dimension) of a 3D array:
X = rand(100, 100, 10)
N = fit(ZScore, X; dims=[1, 2])
normalize!(X, N) # Each [1, 2] slice is normalized independently
all(std(X; dims=[1, 2]) .≈ 1) # trueNormalization methods
Any of these normalizations will work in place of ZScore in the examples above:
Subtract the mean and scale by the standard deviation (aka standardization)
x = 1.5.*randn(100) .+ 0.5
N = fit(ZScore, x)
y = normalize(x, N)
normviz(x, y)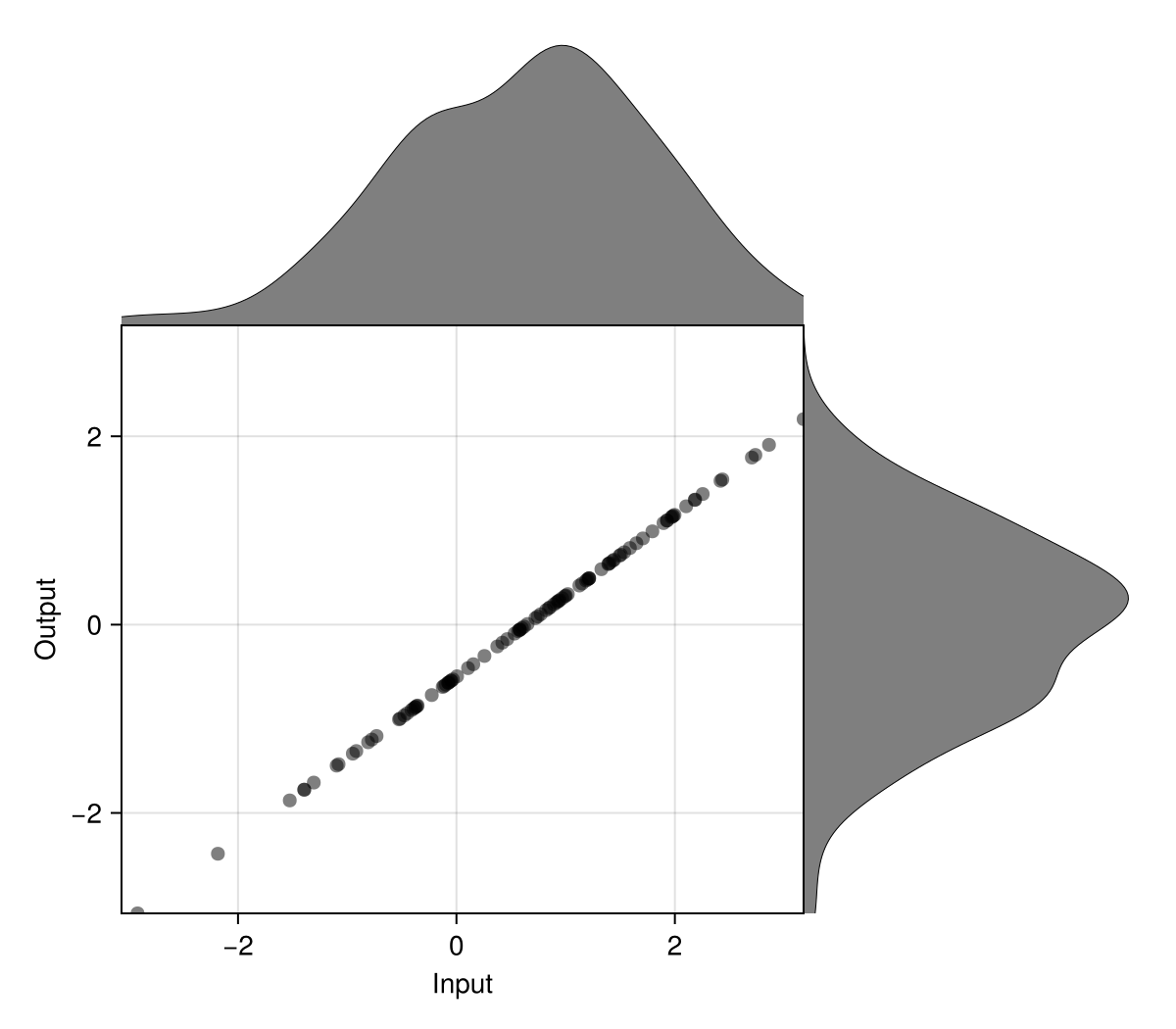
Normalization modifiers
What if the input data contains NaNs or outliers? We provide AbstractModifier types that can wrap an AbstractNormalization to modify its behavior.
Any concrete modifier type Modifier <: AbstractModifier (for example, NaNSafe) can be applied to a concrete normalization type Normalization <:AbstractNormalization:
N = NaNSafe{ZScore} # A combined type with a free `eltype` of `Any`
N = NaNSafe{ZScore{Float64}} # A concrete `eltype` of `Float64`Any AbstractNormalization can be used in the same way as an AbstractModifier.
NaN-safe normalizations
If the input array contains any NaN values, the ordinary normalizations given above will fit with NaN parameters and return NaN arrays. To circumvent this, any normalization can be made 'NaN-safe', meaning it ignores NaN values in the input array, using the NaNSafe modifier.
Robust modifier
The Robust modifier can be used with any AbstractNormalization that has mean and standard deviation parameters. The Robust modifier converts the mean to median and std to iqr/1.35, giving a normalization that is less sensitive to outliers.
Mixed modifier
The Mixed modifier defaults to the behavior of Robust but uses the regular parameters (mean and std) if the iqr is 0.
Properties and traits
The following are common methods defined for all AbstractNormalization subtypes and instances.
Type traits
Normalization.estimators(N::Union{<:AbstractNormalization,Type{<:AbstractNormalization})returns the estimatorsNas a tuple of functionsforward(N::Union{<:AbstractNormalization,Type{<:AbstractNormalization})returns the forward normalization function (e.g.-> for the ZScore)inverse(N::Union{<:AbstractNormalization,Type{<:AbstractNormalization}})returns the inverse normalization function e.g.forward(N)(ps...) |> InverseFunctions.inverseeltype(N::Union{<:AbstractNormalization,Type{<:AbstractNormalization})returns the eltype of the normalization parameters
Concrete properties
Normalization.dims(N::<:AbstractNormalization)returns the dimensions of the normalization. The dimensions are determined bydimsand correspond to the mapped slices of the input array.params(N::<:AbstractNormalization)returns the parameters ofNas a tuple of arrays. The dimensions of arrays are the complement ofdims.isfit(N::<:AbstractNormalization)checks if all parameters are non-empty